A team led by Dr. Chi-Huey Wong has employed saturation mutagenesis to study the influence of peptide sequons in the efficiency of protein N-linked glycosylation, and in collaboration with Dr. Hwai-I Yang, a professional in epidemiology and risk analysis, to establish a multiple statistical model for predicting N-glycosylation. This model predicts the positions and efficiency of protein N-glycosylation by an algorithm that is based on the compositions of 5-residue sequon, and has been successfully applied to other glycoproteins. The study is published in JACS, the Journal of the American Chemical Society, on August 18.
More than 50% of human proteins are glycosylated. The glycans linked to asparagine (N) are called N-linked glycans, which are important co-/post-translational modifications regulating structures and functions of glycoproteins, and closely related to the development of protein drugs. It is known that N-linked glycosylation happens on consensus N-X-S/T sequon [N-X-S/T, N: asparagine, X: any amino acid except for proline, S/T: serine or threonine], but only 65% of the consensus sequon N-X-S/T are detected to have N-linked glycosylation, and without clear data of glycosylation efficiency. Moreover, there are many proteins in databases with predicted N-glycosylation sites awaiting experimental proofs of glycosylation efficiency. Therefore, how to precisely predict the site and efficiency of N-glycosylation is an important topic.
With the rapid development of protein-based biologics, protein glycosylation receives much attention. One of the most famous examples is erythropoietin (EPO)-- enhancing the N-glycosylation of EPO increases the its stability and activity and therefore Amgen adds two more N-glycosylation sites to improve the effectiveness of EPO. Most of the current protein drugs are produced by E. coli or Chinese hamster ovary (CHO) cells. Although CHO cell expression system gives glycosylation that makes it a better option than E. coli for producing the protein drug that requires glycosylation, there is still concern that CHO cell produces non-human glycoforms, and in particular, there is no prediction guide for the efficiency of N-glycosylation. Therefore, prediction of N-glycosylation efficiency for protein engineering to reach better stability and activity is one of the future directions for pharmaceutical industry.
Previous collaboration work of Dr. Wong and Dr. Jeff Kelly (The Scripps Research Institute) indicates that although N-X-S/T are the simplest substrate for oligosaccharyltransferase (OST) to transfer N-linked glycans, the enhanced sequons, which include a nearby aromatic amino acid phenylalanine (F), F-X-N-X-T and F-X-X-N-X-T, are shown to assure effective N-glycosylation (Science 2011, vol 331:571; JACS 2013, vol 135:9877). To further investigate the influence of sequence variation on N-glycosylation efficiency, we used human CD2 adhesion domain (hCD2ad) to screen for the i-2, i-1, i+1 and i+2 residues flanking N at position i by saturation mutagenesis, stepwise screening strategy and applied multi-step statistical calculation to determine the influences of different amino acids in protein N-glycosylation (Figure 1).
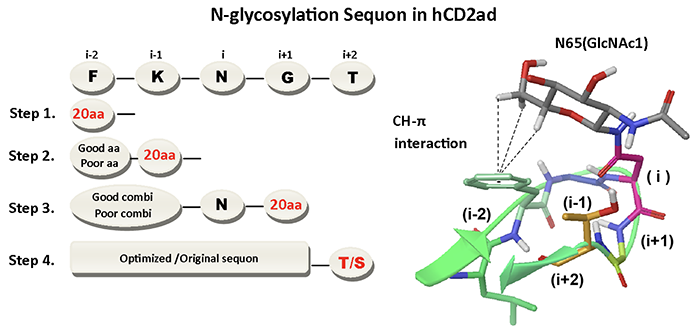 |
| (Fig. 1) |
Figure 2 summarizes the group’s finding that aromatic amino acids [especially tryptophan (W)] and sulfur-containing amino acids at the i-2 position, which is upstream of N residue, improve N-glycosylation efficiency, while positively charged residues such as arginine (R) have a negative effect on N-glycosylation. Thiol- and hydroxyl-containing, and aliphatic amino acids at the i-1 position enhance N-glycosylation efficiency, and cysteine (C) restored the negative effect of R at the i-2 position. Additionally, small amino acids and serine (S) at the i+1 position, which is downstream of N, increase efficiency of N-glycosylation. Based on the level of glycosylation in various sequons, the team collaborated with Dr. Yang to devise an algorithm as a prediction model for the prediction site and N-glycosylation efficiency. The research team introduced optimized sequons to other glycoproteins and further proved that the prediction model achieved very good performance. Moreover, the collaboration work with Dr. Ying-Ta Wu on computer modeling suggested that the high-affinity interaction between the enhanced sequon and an OST subunit (STT3A) was the key to increase glycosylation efficiency. This study provides a better understanding on protein glycosylation through OST, and a prediction guide for controlling glycosylation of proteins at the site of interest.
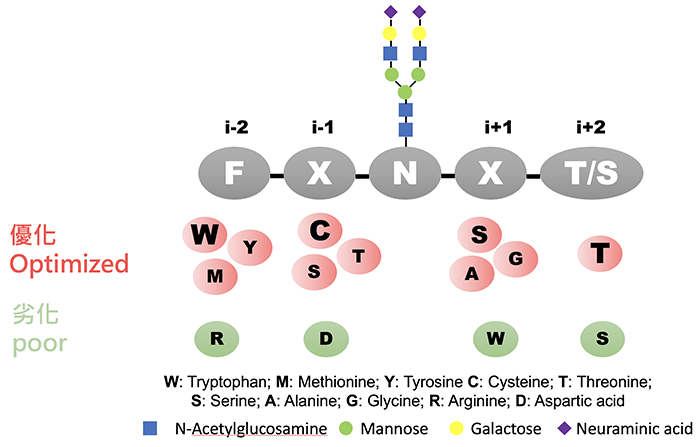 |
| (Fig. 2) |
"From this study, we get to decipher the five amino acid sequons in modulating N-glycosylation efficiency. In addition, this interdisciplinary work between statistics and biochemistry establishes a prediction model that allows for manipulating the N-glycosylation efficiency with a minimal influence on protein structure after replacement of amino acids. This research accomplishment can be applied to increase activity and stability of protein drugs through altering the level of N-glycosylation. This study also gives a hint-- mutation of the amino acids comprising the N-glycosylation sequons may be related to diseases caused by changes of the level of protein N-glycosylation.” commented the team.
This work is published in the Journal of the American Chemical Society in an article entitled “Residues Comprising the Enhanced Aromatic Sequon Influence Protein N-glycosylation Efficiency”. The first author of the publication is Yen-Wen Huang, a Ph.D. graduated from the Institute of Biochemical Sciences, National Taiwan University.
 |
| Team members(from left to right): Dr. Hwai-I Yang, Dr. Yen-Wen Huang, Dr. Tsui-Ling Hsu and Dr. Tzu-Wen Lin。 |


