Peptidoglycan is a major component of the bacterial cell wall and essential for bacteria to survive. Bacterial transpeptidase and transglycosylase on the cell surface are the two key enzymes responsible for making peptidoglycan. Many current antibiotics have been developed to target the transpeptidase, but the problem of antibiotic resistance has arisen and caused a major threat in treating bacterial infection, such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE). When a thorough examination is done toward both Gram positive and Gram negative bacteria, including MRSA, VRE, and drug-resistant bacteria of Acinetobacter baumannii, Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa, and Mycobacterium tuberculosis, it appeared that none has a gene mutation in the transglycosylase area. The transglycosylase has been considered to be another excellent target, but no antibiotics have been developed to target this enzyme.
The research team at the Genomics Research Center, Academia Sinica has solved the structure of Staphylococcus aureus transglycosylase, together with its substrate lipid II analog, to provide not only a basis for understanding the mechanism of how the enzyme transglycoslase takes lipid II and makes it into peptidoglycan, but also a foundation for new antibiotic design against transglycosyase.
From the study, the team proposed how the enzyme transglycosylase dimerizes two lipid II molecules into one lipid IV molecule in atomic details (Fig. 1).
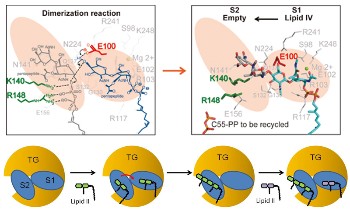 |
| Figure 1 |
This work provides the atomic information for understanding the key step of how the cell wall of bacteria, peptidoglycan, is built, and paves the way for structure-based antibiotic design against transglycosylase. The research is co-led by President Chi-Huey Wong and Dr. Che Alex Ma, and is published online on PNAS website on April 9th, 2012, and Ms. Chia-Ying Huang is the first author.


