Rubik’s Cube is one of the most popular 3D combinatorial games that captivates the world with its amazing complexity hidden behind the deceptively simplistic six bright colors. Remarkably, the most accomplished players can solve a completely scrambled Rubik’s Cube in less than twelve seconds. Few people, however, know that there is an equally complex Rubik’s Cube-like molecular machine in the cell. This machine is called “Spliceosome”, which must be sequentially remodeled or tweaked, just like the Rubik’s Cube, before it can be properly functioning in the gene expression pathway.
During gene expression, the genetic information stored in the DNA molecule (think, computer hard drive) must first be transcribed (or copied) into messenger RNA (mRNA), which is then exported into cytoplasm for translation into protein molecule. But, this seemingly straightforward paradigm is in fact quite complicated.
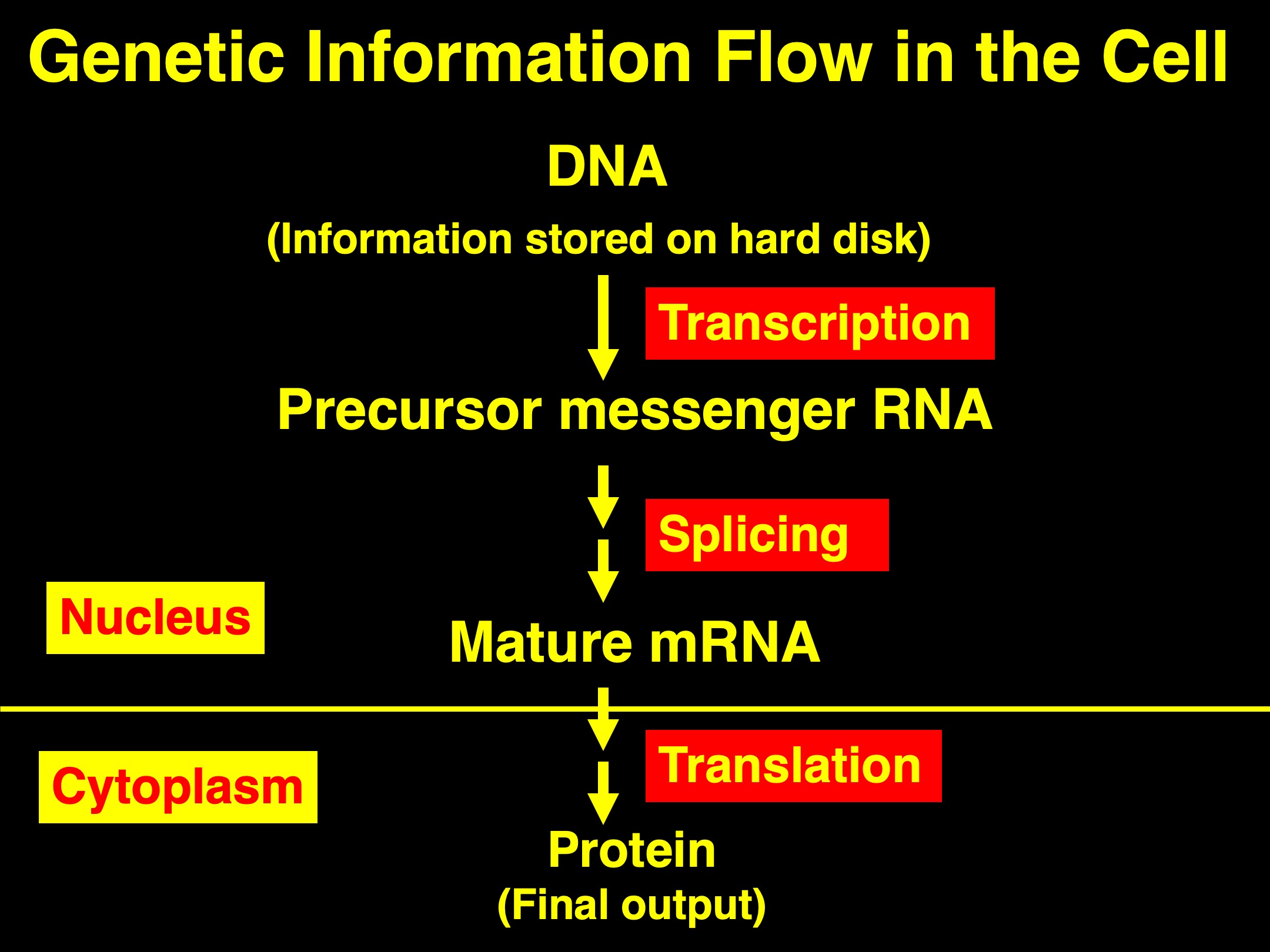
Most eukaryotic genes are discontinuous on the DNA template. That is, the coding information of a gene is split into a series of exons (coding region) and introns (non-coding region). To express any given gene, the genetic information carried on the DNA template needs to be first transcribed into precursor messenger RNAs (pre-mRNAs), on which both exons and introns are still preserved. The next step for gene expression is then to cut out, at the pre-mRNA level, the non-coding introns and stitched the coding exons together. This process is called “Splicing”, which results in matured mRNAs. Export of matured mRNAs from the nucleus, where transcription and splicing both occur, into cytoplasm will then allow the matured mRNA be translated into proteins, the final products of the gene expression pathway.
Now, imagine the challenge of the splicing process faces. For example, the non-coding intron may have a length stretching over 100 kilobases (say, from Taipei to Kaohsiung, metaphorically speaking). So how could it be possible that a cell can recognize the two ends of intron and cut the intron out precisely? No wonder, this splicing process was found to be achieved by a ~200-part molecular machine, called spliceosome. Spliceosome is an incredibly precise machinery, which must undergo a stepwise of binding and conformational rearrangements. Through remodeling, it permits the checking and rechecking of RNA sequences before the chemical reaction is allowed to proceed. This series of remodeling is performed by RNA helicases. But exactly how RNA helicases play the cellular Rubik’s Cube, i.e., the spliceosome, is a long-standing mystery.
Enter Professor Tien-Hsien Chang’s research group. For years, Dr. Chang has been trying to unravel this mystery using Prp28, a spliceosomal RNA helicase, as a model system.
“I wanted to know how an enzyme does its job on a very big molecular complex,” said Dr. Chang, “which means how this Rubik’s cube gets played by these RNA helicases”.
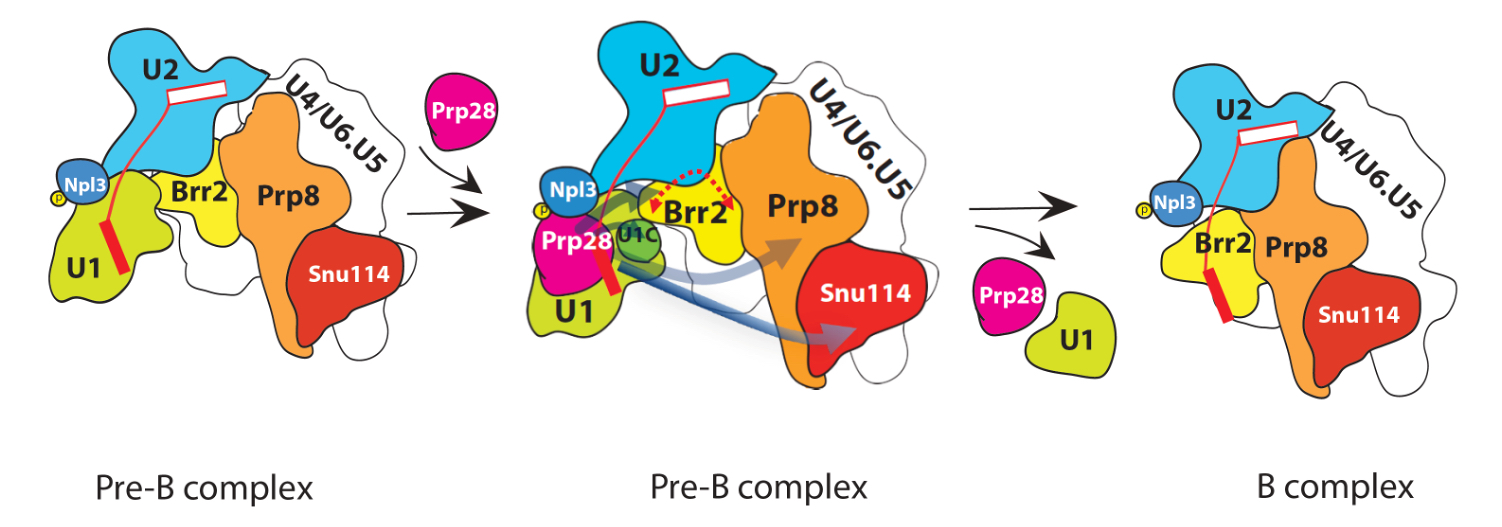
Some years ago, Prof. Chang’s group demonstrated Prp28, an RNA helicase, is responsible to remove U1 snRNP from a fully built spliceosome. But mechanistically how that is done remains unknown. Now, through more than a decade of effort, Dr. Chang’s group finally cracks the toughest nut.
“This is a very difficult problem, because these enzymes generally come in, and do their job and then leave quickly. So, it’s very hard to capture how exactly they work on this molecular complex.” said Dr. Chang.
To capture the transient interactions (lasting for only few seconds) of Prp28 with the spliceosome to get the job done, they adapted a highly ingenious method. They placed cross-linking reagent (think, “instant glue”) at 42 different positions respectively on Prp28, which was done in a one-by-one painstaking manner. When the instant-glue modified Prp28 is introduced into splicing system in the test tube and ultraviolet light (UV) is shined on the reaction, the “instant glue” will capture Prp28-near-by components within the complex spliceosome. Using this method, they systematically mapped out the components that Prp28 physically interacts with and the specific time points that these interactions take place. They then comprehensively verified the biological meaning of each interaction by sophisticated genetics and biochemistry.
It took Dr. Fu-Lung Yeh in Dr. Chang’s lab two years to begin to detect the first faint signals and nearly another decade to complete this series of highly sophisticated experiments. Most critically, Dr. Yeh identified a co-factor of Prp28, i.e., the phosphorylated form of Npl3, an RNA binding protein.
It turns out that phosphorylation of Npl3 attracts or recruits Prp28 to the right place at the right time within the spliceosome. It is there at the right time Prp28’s ATPase activity was unleased through its interaction with phosphorylated Npl3. Hydrolysis of ATP by Prp28 thus provides the needed energy to eject U1 snRNP, so as to complete the first step of spliceosome remodeling. Without completing this step, spliceosome cannot move forward for the splicing reaction. And, of course, without splicing, cells shall die.
Splicing is an indispensable step of gene expression. Defects or inaccuracies during the assembly or the rearrangement steps have been linked to diverse hereditary diseases. The results of this research shall serve as a conceptual and technical paradigm for mechanistic understanding the functions of other RNA helicases on the spliceosome and on many RNA-related machines as well in the cell.
“After a marathon-like seemingly no-end-in-sight struggle to cross the finishing line, I am delighted that I never gave it up and kept it going until I call it a victory” said Dr. Chang, “If you’re truly obsessed with a Herculean challenge, then you are determined to throw your life with blood and sweat to overcome all the mounting difficulties. And I must say that being able to solve one of the most difficult problems in science does give me tons of satisfaction”.
The paper can be read online at: Yeh, FL., Chang, SL., Ahmed, G.R. et al. Activation of Prp28 ATPase by phosphorylated Npl3 at a critical step of spliceosome remodeling. Nat Commun 12, 3082 (2021). https://doi.org/10.1038/s41467-021-23459-4


